Aortic valve reconstruction in non-contrast computed tomography: a new approach
Accurate aortic valve reconstruction and segmentation in computed tomography (CT) is crucial for diagnosing heart disease. This information is essential for planning effective interventions such as surgical and percutaneous procedures. However, non-contrast CT scans often present challenges due to the AV’s low visibility.
Moreover, non-contrast AV segmentation shows promise for radiotherapy planning near the heart, where radiation exposure to specific cardiac substructures increases cardiotoxicity risk.
Manual segmentation, a traditional approach, is both time-consuming and prone to errors.[1]
The challenge of aortic valve reconstruction and segmentation in non-contrast scans
Non-contrast tomography struggles to differentiate the aorta from nearby organs and tissues due to their similar radiological densities. This makes accurately locating the valve challenging. Consequently, it is very difficult to identify calcifications belonging to that region, which is crucial for assessing the severity of aortic stenosis, a key diagnostic factor.
In summary, while non-contrast CT excels at detecting calcifications, it faces challenges in distinguishing vascular structures due to limited contrast. Contrast-enhanced CT offers superior visualization of vessels but may obscure calcifications. Both techniques provide complementary information for cardiac imaging. However, relying exclusively on non contrast CT is essential for calcium scoring and has great potential, especially for patients who cannot receive contrast agents.[2]
A novel approach to AV segmentation
Segmentation of AV is essential for accurately calculating the Agatston score, a critical indicator in assessing coronary artery disease.
To address the above challenges, experts at Graylight Imaging have proposed a semi-automatic method for generating ground truth (GT) data based on image registration, which allows for effective training of machine learning (ML) segmentation models in non-contrast CT. Following this approach, in a weakly supervised learning process, we train neural network models that can accurately segment the AV solely based on non-contrast scans. This approach makes the segmentation of the aortic root and valve more accessible for clinical applications. We also present a novel approach to evaluating segmentation accuracy, based on rigid registration of segmented masks in contrast and non-contrast images. [3]
Scalable model for aortic root segmentation
In our study, we successfully developed a scalable ML model for segmenting the aortic root in non-contrast images. Moreover, employing a weakly supervised learning technique described above, we have created a tool that can precisely locate the AV with an accuracy of 0.8 mm, which we estimated based on a specialized version of the iterative closest point (ICP) algorithm. Our solution represents a significant advancement in the field of medical image analysis.[4]
Scalability and efficiency
The scalability of our method is demonstrated by its ability to almost fully automate the processing of potentially unlimited datasets without significantly increasing the workload for human operators. Initially, our process relies on a conventional registration method between contrast and non-contrast images. Subsequently, human annotator intervention is required, but this involvement is limited to simple tasks of approving or rejecting registration results. Typically, this stage requires about 1 minute per scan for an experienced annotator. Furthermore, the evaluation technique operates entirely autonomously, eliminating the need for manual intervention.[5]
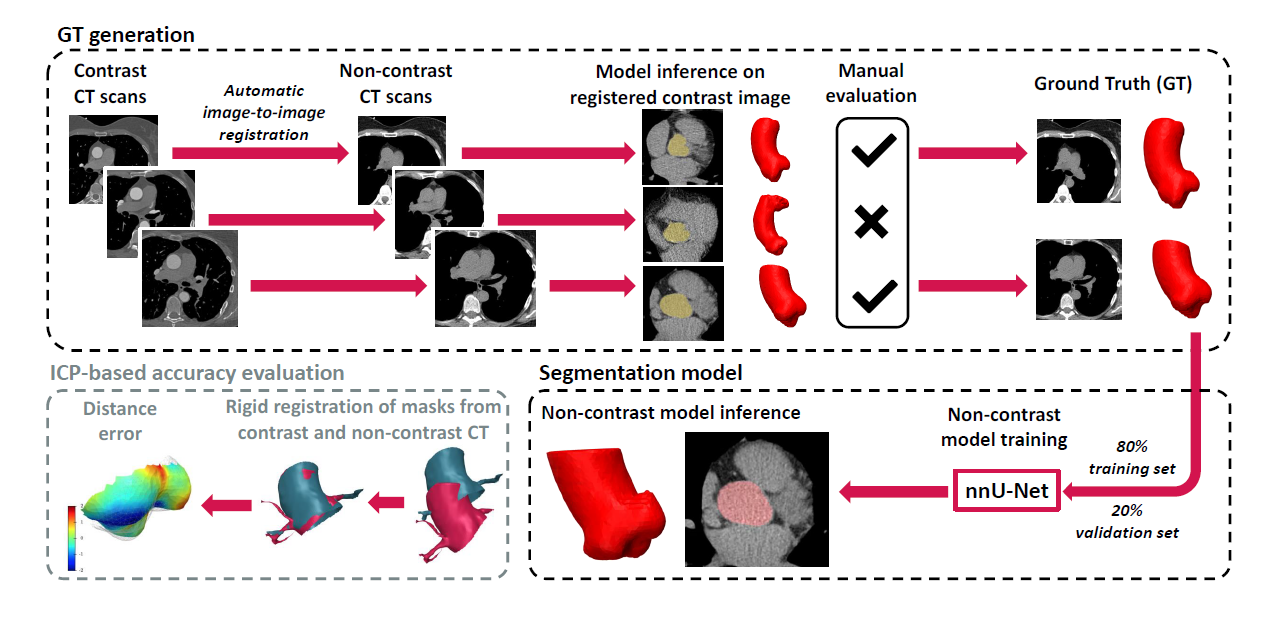
Main steps of the framework for training and evaluation of the aortic root
segmentation neural network based on non-contrast CT scans.
Generating training data
Primarily, to generate input masks used in subsequent image registration steps, we segment aortic roots in contrast-enhanced computed tomography scans. Due to the good visibility of the aorta, including the aortic root, in this modality, both manual delineation based on standard procedures and automatic segmentation using state-of-the-art deep learning models are possible.[6]
Aortic root segmentation in non-contrast CT images
In the next step, we perform image registration of 268 contrast-enhanced CT scans to their non-contrast counterparts, based on the standard Greedy framework. A semi-supervised deep learning technique, based on the previously described registration and annotation process, allows us to create a model capable of segmenting the aortic root directly from non-contrast images.[7]
Evaluation of model accuracy
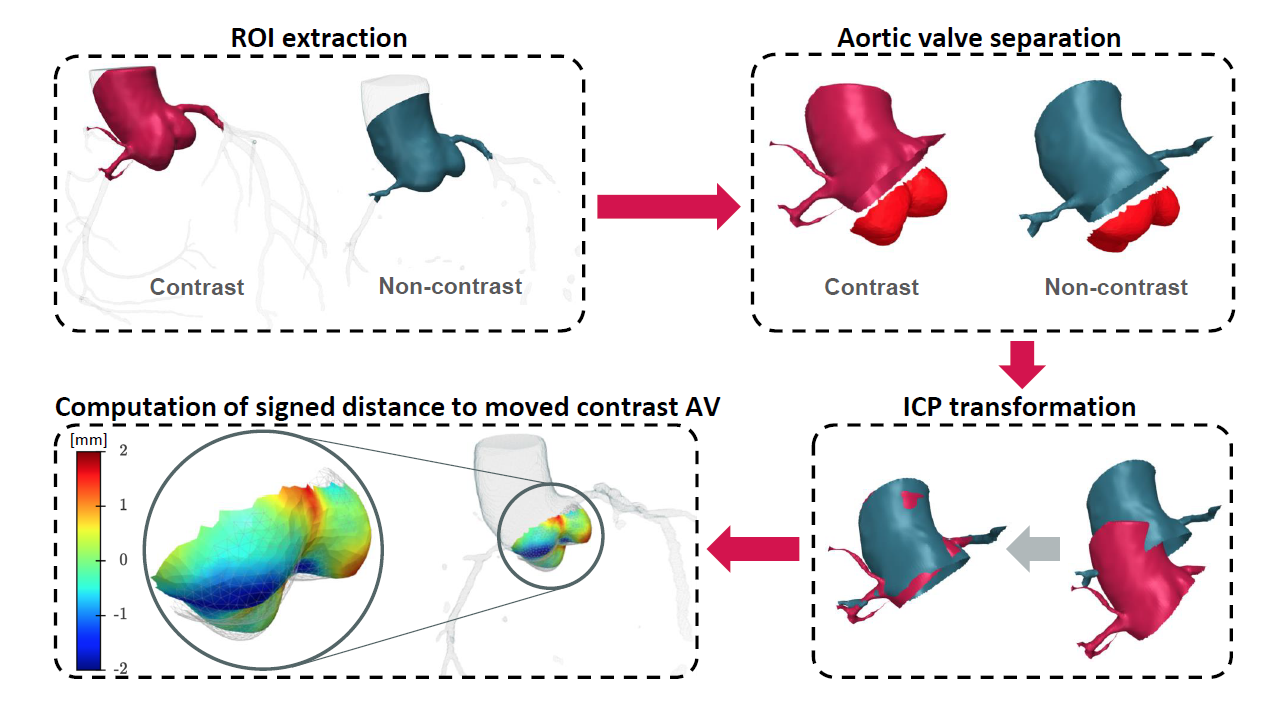
The procedure of evaluation of aortic valve accuracy
Qualitative inspections of the geometry of the aorta created by the model in non-contrast images suggest that it not only reproduces the aortic root in detail but also extrapolates the shape and position of the AV. To quantitatively assess the accuracy of the model, we applied the ICP-based alignment procedure, comparing the results of our model with manually segmented GT data.[8]
Cutting edge machine learning techniques in aortic valve reconstruction: a summary
Our research demonstrates that accurate segmentation of the aortic valve and aortic root in non-contrast scans is possible thanks to modern ML techniques. The obtained results suggest that the model can be successfully applied in clinical practice, especially in the assessment of coronary artery disease and radiotherapy planning. Thanks to our method, the accuracy of AV segmentation reached a mean error of 0.8 mm, making it a valuable tool in everyday diagnostics. This approach offers new perspectives in image diagnostics, enabling precise segmentation even in difficult cases where traditional methods have failed.[9]
References:
[1]-[9] Bujny, M., Jesionek, K., Nalepa, J., Bartczak, T., Miszalski-Jamka, K., Kostur, M. (2024). Seeing the Invisible: On Aortic Valve Reconstruction in Non-contrast CT. In: Linguraru, M.G., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2024. MICCAI 2024. Lecture Notes in Computer Science, vol 15009. Springer, Cham, https://link.springer.com/chapter/10.1007/978-3-031-72114-4_55.